process¶
peak_alignment¶
-
dimspy.process.peak_alignment.align_peaks(peaks: Sequence[dimspy.models.peaklist.PeakList], ppm: float = 2.0, block_size: int = 5000, fixed_block: bool = True, edge_extend: Union[int, float] = 10, ncpus: Optional[int] = None)[source]¶ Cluster and align peaklists into a peak matrix.
- Parameters
peaks – List of peaklists for alignment
ppm – The hierarchical clustering cutting height, i.e., ppm range for each aligned mz value. Default = 2.0
block_size – number peaks in each centre clustering block. This can be a exact or approximate number depends on the fixed_block parameter. Default = 5000
fixed_block – Whether the blocks contain fixed number of peaks. Default = True
edge_extend – Ppm range for the edge blocks. Default = 10
ncpus – Number of CPUs for parallel clustering. Default = None, indicating using as many as possible
- Return type
PeakMatrix object
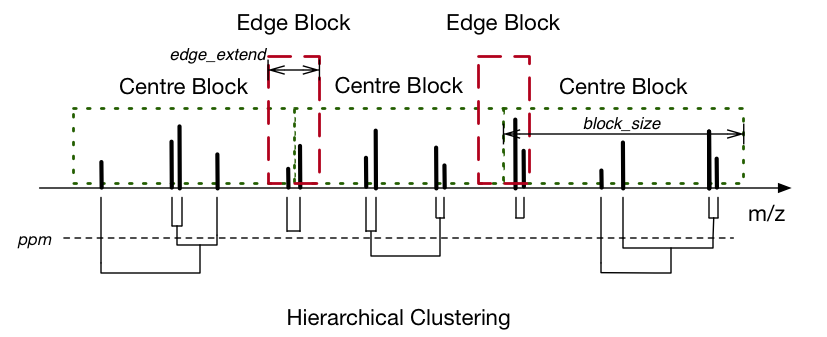
This function uses hierarchical clustering to align the mz values of the input peaklists. The alignment “width” is decided by the parameter of ppm. Due to a large number of peaks, this function splits them into blocks with fixed or approximate length, and clusters in a parallel manner on multiple CPUs. When running, the edge blocks are clustered first to prevent separating the same peak into two adjacent centre blocks. The size of the edge blocks is decided by edge_extend. The clustering of centre blocks is conducted afterwards.
After merging the clustering results, all the attributes (mz, intensity, snr, etc.) are aligned into matrix accordingly. If multiple peaks from the same sample are clustered into one mz value, their attributes are averaged (for real value attributes e.g. mz and intensity) or concatenated (string, unicode, or bool attributes). The flag attributes are ignored. The number of these overlapping peaks is recorded in a new intra_count attribute matrix.
peak_filters¶
-
dimspy.process.peak_filters.filter_attr(pl: dimspy.models.peaklist.PeakList, attr_name: str, max_threshold: Optional[Union[int, float]] = None, min_threshold: [<class 'int'>, <class 'float'>, None] = None, flag_name: Optional[str] = None, flag_index: Optional[int] = None)[source]¶ Peaklist attribute values filter.
- Parameters
pl – The target peaklist
attr_name – Name of the target attribute
max_threshold – Maximum threshold. A peak will be unflagged if the value of it’s attr_name is larger than the threshold. Default = None, indicating no threshold
min_threshold – Minimum threshold. A peak will be unflagged if the value of it’s attr_name is smaller than the threshold. Default = None, indicating no threshold
flag_name – Name of the new flag attribute. Default = None, indicating using attr_name + ‘_flag’
flag_index – Index of the new flag to be inserted into the peaklist. Default = None
- Return type
PeakList object
This filter accepts real value attributes only.
-
dimspy.process.peak_filters.filter_ringing(pl: dimspy.models.peaklist.PeakList, threshold: float, bin_size: Union[int, float] = 1.0, flag_name: str = 'ringing_flag', flag_index: Optional[int] = None)[source]¶ Peaklist ringing filter.
- Parameters
pl – The target peaklist
threshold – Intensity threshold ratio
bin_size – size of the mz chunk for intensity filtering. Default = 1.0 ppm
flag_name – Name of the new flag attribute. Default = ‘ringing_flag’
flag_index – Index of the new flag to be inserted into the peaklist. Default = None
- Return type
PeakList object
This filter will split the mz values into bin_size chunks, and search the highest intensity value for each chunk. All other peaks, if it’s intensity is smaller than threshold x the highest intensity in that chunk, will be unflagged.
-
dimspy.process.peak_filters.filter_mz_ranges(pl: dimspy.models.peaklist.PeakList, mz_ranges: Sequence[Tuple[float, float]], flag_name: str = 'mz_ranges_flag', flagged_only: bool = False, flag_index: Optional[int] = None)[source]¶ Peaklist mz range filter.
- Parameters
pl – The target peaklist
mz_ranges – The mz ranges to remove. Must be in the format of [(mz_min1, mz_max2), (mz_min2, mz_max2), …]
flag_name – Name of the new flag attribute. Default = ‘mz_range_remove_flag’
flag_index – Index of the new flag to be inserted into the peaklist. Default = None
- Return type
This filter will remove all the peaks whose mz values are within any of the ranges in the mz_remove_rngs.
-
dimspy.process.peak_filters.filter_rsd(pm: dimspy.models.peak_matrix.PeakMatrix, rsd_threshold: Union[int, float], qc_tag: Any, on_attr: str = 'intensity', flag_name: str = 'rsd_flag')[source]¶ PeakMatrix RSD filter.
- Parameters
pm – The target peak matrix
rsd_threshold – Threshold of the RSD of the QC samples
qc_tag – Tag (label) to unmask qc samples
on_attr – Calculate RSD on given attribute. Default = “intensity”
flag_name – Name of the new flag. Default = ‘rsd_flag’
- Return type
This filter will calculate the RSD values of the QC samples. A peak with a QC RSD value larger than the threshold will be unflagged.
-
dimspy.process.peak_filters.filter_fraction(pm: dimspy.models.peak_matrix.PeakMatrix, fraction_threshold: float, within_classes: bool = False, class_tag_type: Any = None, flag_name: str = 'fraction_flag')[source]¶ PeakMatrix fraction filter.
- Parameters
pm – The target peak matrix
fraction_threshold – Threshold of the sample fractions
within_classes – Whether to calculate the fraction array within each class. Default = False
class_tag_type – Tag type to unmask samples within the same class (e.g. “classLabel”). Default = None
flag_name – Name of the new flag. Default = ‘fraction_flag’
- Return type
PeakMatrix object
This filter will calculate the fraction array over all samples or within each class (based on class_tag_type). The peaks with a fraction value smaller than the threshold will be unflagged.
-
dimspy.process.peak_filters.filter_blank_peaks(pm: dimspy.models.peak_matrix.PeakMatrix, blank_tag: Any, fraction_threshold: Union[int, float] = 1, fold_threshold: Union[int, float] = 1, method: str = 'mean', rm_blanks: bool = True, flag_name: str = 'blank_flag')[source]¶ PeakMatrix blank filter.
- Parameters
pm – The target peak matrix
blank_tag – Tag (label) to mask blank samples. e.g Tag(“blank”, “classLabel”)
fraction_threshold – Threshold of the sample fractions. Default = 1
fold_threshold – Threshold of the blank sample intensity folds. Default = 1
method – Method to calculate blank sample intensity array. Valid values include ‘mean’, ‘median’, and ‘max’. Default = ‘mean’
rm_blanks – Whether to remove (not mask) blank samples after filtering
flag_name – Name of the new flag. Default = ‘blank_flag’
- Return type
PeakMatrix object
This filter will calculate the intensity array of the blanks using the “method”, and compare with the intensities of the other samples. If fraction_threshold% of the intensity values of a peak are smaller than the blank intensities x fold_threshold, this peak will be unflagged.
scan_processing¶
-
dimspy.process.replicate_processing.remove_edges(pls_sd: Dict)[source]¶ Removes overlapping m/z regions of adjacent (SIM) windows / scan events.
- Parameters
pls_sd – List of peaklist objects
- Returns
List of peaklist objects
-
dimspy.process.replicate_processing.read_scans(fn: str, function_noise: str, min_scans: int = 1, filter_scan_events: Dict = None)[source]¶ Read, filter, group and sort scans based on the header / filter string Helper function for ‘process_scans (tools module)’
- Parameters
fn – Path to the .mzml or .raw file
function_noise –
Function to calculate the noise from each scan. The following options are available:
median - the median of all peak intensities within a given scan is used as the noise value.
mean - the unweighted mean average of all peak intensities within a given scan is used as the noise value.
mad (Mean Absolute Deviation) - the noise value is set as the mean of the absolute differences between peak intensities and the mean peak intensity (calculated across all peak intensities within a given scan).
noise_packets - the noise value is calculated using the proprietary algorithms contained in Thermo Fisher Scientific’s msFileReader library. This option should only be applied when you are processing .RAW files.
min_scans – Minimum number of scans required for each m/z window or event within a raw/mzML data file.
filter_scan_events –
Include or exclude specific scan events, by default all ALL scan events will be included. To include or exclude specific scan events use the following format of a dictionary.
>>> {"include":[[100, 300, "sim"]]} or {"include":[[100, 1000, "full"]]}
- Returns
List of peaklist objects
-
dimspy.process.replicate_processing.average_replicate_scans(name: str, pls: Sequence[dimspy.models.peaklist.PeakList], ppm: float = 2.0, min_fraction: float = 0.8, rsd_thres: float = 30.0, rsd_on: str = 'intensity', block_size: int = 5000, ncpus: int = None)[source]¶ Align, filter and average replicate scans/peaklist Helper function for ‘process_scans (tools module)’
- Parameters
name – Name average peaklist
pls – List of peaklists
ppm – Maximum tolerated m/z deviation in parts per million.
min_fraction – A numerical value from 0 to 1 that specifies the minimum proportion of scans a given mass spectral peak must be detected in, in order for it to be kept in the output peaklist. Here, scans refers to replicates of the same scan event type, i.e. if set to 0.33, then a peak would need to be detected in at least 1 of the 3 replicates of a given scan event type.
rsd_thres – Relative standard deviation threshold - A numerical value equal-to or greater-than 0. If greater than 0, then peaks whose intensity values have a percent relative standard deviation (otherwise termed the percent coefficient of variation) greater-than this value are excluded from the output peaklist.
rsd_on – Intensity or SNR
block_size – Number peaks in each centre clustering block.
ncpus – Number of CPUs for parallel clustering. Default = None, indicating using all CPUs that are available
- Returns
List of peaklists
-
dimspy.process.replicate_processing.average_replicate_peaklists(pls: Sequence[dimspy.models.peaklist.PeakList], ppm: float, min_peaks: int, rsd_thres: float = None, block_size: int = 5000, ncpus: int = None)[source]¶ Align, filter and average replicate peaklists. Helper function for ‘replicate_filter (tools module)’
- Parameters
pls – List of peaklists
ppm – Maximum tolerated m/z deviation in parts per million.
min_peaks – Minimum number of technical replicates (i.e. peaklists) a peak has to be present in.
rsd_thres – Relative standard deviation threshold - A numerical value equal-to or greater-than 0. If greater than 0, then peaks whose intensity values have a percent relative standard deviation (otherwise termed the percent coefficient of variation) greater-than this value are excluded from the output peaklist.
block_size – Number peaks in each centre clustering block.
ncpus – Number of CPUs for parallel clustering. Default = None, indicating using all CPUs that are available
- Returns
List of peaklists
-
dimspy.process.replicate_processing.join_peaklists(name: str, pls: Sequence[dimspy.models.peaklist.PeakList])[source]¶ Join/Merge peaklists (i.e. windows) with different m/z ranges. Helper function for ‘process_scans (tools module)’
- Parameters
name – Name newly created joined/merged peaklist
pls – List of peaklists
- Returns
Peaklist